
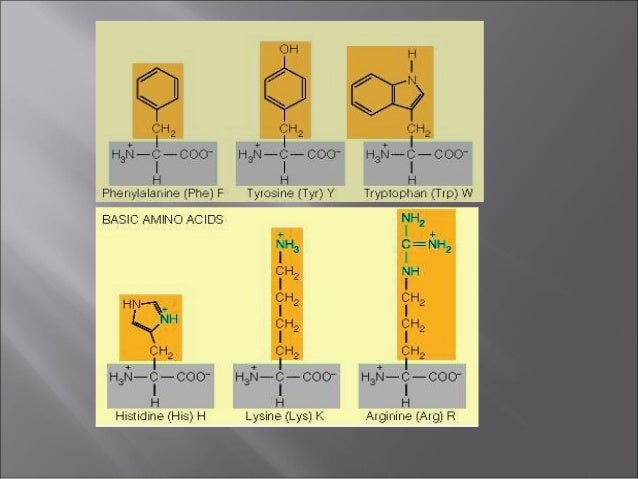
These findings elucidate the mechanism of an intramembrane chaperone and suggest a new framework for multipass membrane protein biogenesis at the endoplasmic reticulum. d, Space-filling models showing the multipass translocon’s highly conserved interior surface that. Accordingly, biogenesis of several multipass proteins was unimpeded by inhibitors of the Sec61 lateral gate. Key structural elements, including the density assigned to TMD3 of the substrate, are indicated. Detection of multiple TMDs in this cavity after their emergence from the ribosome suggests that multipass proteins insert and fold behind Sec61. Instead, Asterix binds to and redirects the substrate to a location behind Sec61, where the PAT complex contributes to a multipass translocon surrounding a semi-enclosed, lipid-filled cavity 11. Here, biochemical and structural analysis of intermediates during multipass protein biogenesis showed that the nascent chain is not engaged with Sec61, which is occluded and latched closed by CCDC47. The PAT complex, an intramembrane chaperone comprising Asterix and CCDC47, engages early TMDs of multipass proteins to promote their biogenesis by an unknown mechanism 10. Thus, the EMC ena- bles the biogenesis and folding of a subset of multipass membrane proteins which present chal- lenges for the canonical membrane protein. The prevailing model posits that each transmembrane domain (TMD) of a multipass protein successively passes into the lipid bilayer through a front-side lateral gate of the Sec61 protein translocation channel 3-9. For example, Claudin18.2, CD20, and CD133 have. How multipass proteins are co-translationally inserted and folded at the endoplasmic reticulum is not well understood 2. Multi-pass transmembrane proteins span the cell membrane multiple times forming multiple extracellular domains. Multipass membrane proteins play numerous roles in biology and include receptors, transporters, ion channels and enzymes 1,2. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins 2018. Diversity, Equity, Inclusion, and Access.Citation, Usage, Privacy Policies, Logo.Our results reveal universal sequence-structure principles governing the complex anatomy and plasticity of multipass membrane proteins that may guide de novo structure prediction. Biologically Interesting Molecule Reference Dictionary (BIRD) Interacting TMHs' topology and local protein conformational flexibility were remarkably well predicted in a blinded fashion from the identified binding-hotspot motifs.


 0 kommentar(er)
0 kommentar(er)
